com_eco
Community Ecology Basics
Nilanjan Chatterjee
Nestedness
Nestedness is a measure of structure in an ecological system. There are different measures for nestedness. One common measure is calculating the temperature. We will use that measure to calculate nestedness here. The value ranges from 0-100 where 0 stands for perfectly nested community and 100 stands for random community
Vegan is one the most important library for analysis of community dataset. The most common type of community data is sites x species data. We would uploda a dummy dataset in the system and leran how to calculate nestedness.
Import the data in R
library(vegan)
setwd("C:/Users/HP/Documents/com_eco")
bhagbird <-read.csv("allbird_38.csv")
head(bhagbird)
Calculating nestedness
Nestedness can be calculated by various methods (Almeida-Neto et al., 2008, Baselga 2012). The references of the methods are given at the bottom of the page. Here I am showing a simple method to calcualte nestedness. This measure is based on the matrix temperature which is defined as the sum of “surprises” in arranged matrix. For the calculation, the site x species matrix is arranged based on sites with higher number of species and species present at maximum sites.
nesttry<-nestedtemp(bhagbird[,2:15])
nesttry
## nestedness temperature: 17.74614
## with matrix fill 0.4450549Including Plots
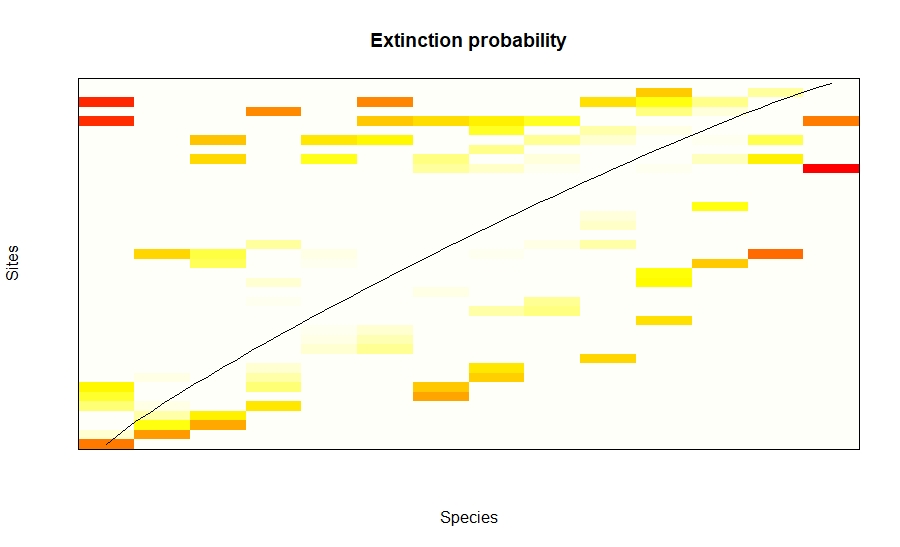
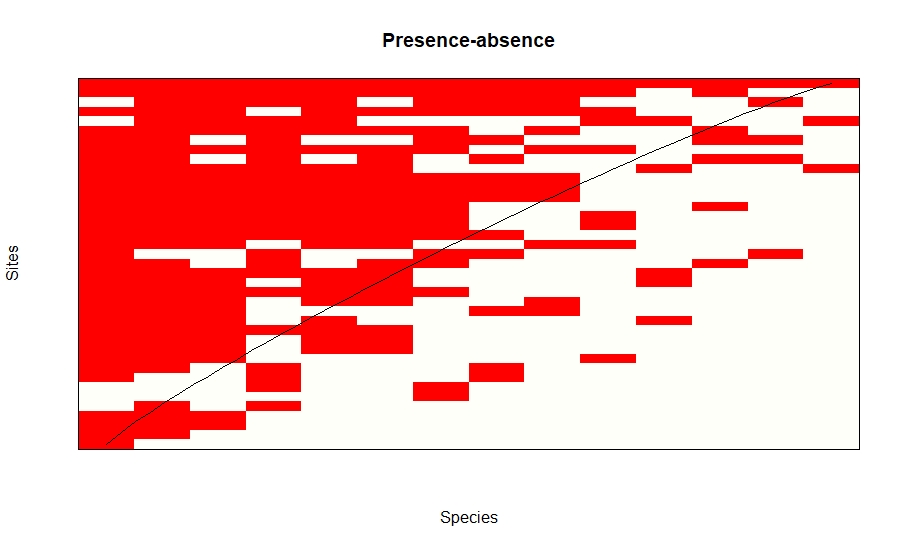
The plots for nestedness temperature is given by these plots. Darker the colour means higher the probability of the species extinction from the site.
plot(nesttry, xlab = "Species", ylab="Sites",main="Extinction probability")
plot(nesttry, kind="incidence",xlab = "Species", ylab="Sites",main="Presence-absence")
Beta-diversity
Similar to nestedness, there is another measure to understand the turnover of species in different community. The measure is known as Beta-diversity. We would use the package betapart for calculation of beta-diversity
library(betapart)
spr_14 <-read.csv("spring_14.csv")
head(spr_14)
## X X.1 Phalacrocorax_niger Vanellus_duvaucelii
## 1 Triveni ghat 2014 1 1
## 2 Ghat 1 2014 1 1
## 3 Ghat 2 2014 1 0
## 4 Ramjhula 2014 0 1
## 5 Phoolchatti main river 2014 1 0
## 6 Phoolchatti side stream 2014 1 0
## Ibidorhyncha_struthersii Actitis_hypoleucos Cinclus_pallasii
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 0 0 1
## Enicurus_scouleri Enicurus_maculatus Myophonus_caeruleus
## 1 0 0 0
## 2 0 0 0
## 3 0 0 0
## 4 0 0 0
## 5 0 0 0
## 6 1 0 0
## Chaimarrornis_leucocephalus Rhyacornis_fuliginosa
## 1 0 0
## 2 0 0
## 3 0 0
## 4 0 0
## 5 0 0
## 6 0 0
## Motacilla_madaraspatensis Motacilla_cinerea Motacilla_alba
## 1 2 0 0
## 2 1 0 0
## 3 1 0 0
## 4 4 0 0
## 5 1 0 0
## 6 1 0 0
## Megaceryle_lugubris Halcyon_smyrnensis Alcedo_atthis
## 1 1 1 2
## 2 0 1 0
## 3 1 0 1
## 4 1 1 0
## 5 1 0 0
## 6 3 1 3Beta-diversity calculation using abundance data
beta.try <-beta.pair.abund(spr_14[,3:18])
beta_abd <- as.matrix(beta.try$beta.bray)
beta_abd[upper.tri(beta_abd, diag = FALSE)] <- " "
head(beta_abd)
## 1 2 3
## 1 "0" "" ""
## 2 "0.333333333333333" "0" ""
## 3 "0.333333333333333" "0.5" "0"
## 4 "0.333333333333333" "0.454545454545455" "0.636363636363636"
## 5 "0.454545454545455" "0.428571428571429" "0.142857142857143"
## 6 "0.368421052631579" "0.6" "0.466666666666667"
## 4 5 6 7 8 9 10 11 12 13 14 15
## 1 "" "" "" "" "" "" "" "" "" "" "" ""
## 2 "" "" "" "" "" "" "" "" "" "" "" ""
## 3 "" "" "" "" "" "" "" "" "" "" "" ""
## 4 "0" "" "" "" "" "" "" "" "" "" "" ""
## 5 "0.6" "0" "" "" "" "" "" "" "" "" "" ""
## 6 "0.666666666666667" "0.571428571428571" "0" "" "" "" "" "" "" "" "" ""
## 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39
## 1 "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" ""
## 2 "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" ""
## 3 "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" ""
## 4 "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" ""
## 5 "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" ""
## 6 "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" "" ""
## 40 41 42 43
## 1 "" "" "" ""
## 2 "" "" "" ""
## 3 "" "" "" ""
## 4 "" "" "" ""
## 5 "" "" "" ""
## 6 "" "" "" ""Beta-diversity calculation using presence-absence data
spr_14_pa <-spr_14
spr_14_pa[spr_14_pa > 0] <-1
class(spr_14_pa)
beta.try.pa <-beta.pair(spr_14_pa[,3:18])
beta_pa <- as.matrix(beta.try.pa$beta.sor)
beta_pa[upper.tri(beta_pa, diag = FALSE)] <- " "
head(beta_pa)
Reference
Almeida-Neto, M., Guimaries, P., Guimaries, P.R., Loyola, R.D. & Ulrich, W. (2008). A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos 117, 1227-1239.
Almeida-Neto, M. & Ulrich, W. (2011). A straightforward computational approach for measuring nestedness using quantitative matrices. Env. Mod. Software 26, 173-178.
Atmar, W. & Patterson, B.D. (1993). The measurement of order and disorder in the distribution of species in fragmented habitat. Oecologia 96, 373-382.
Baselga, A. (2012). The relationship between species replacement, dissimilarity derived from nestedness, and nestedness. Global Ecol. Biogeogr. 21, 1223-1232.
Brualdi, R.A. & Sanderson, J.G. (1999). Nested species subsets, gaps, and discrepancy. Oecologia 119, 256-264.
Patterson, B.D. & Atmar, W. (1986). Nested subsets and the structure of insular mammalian faunas and archipelagos. Biol. J. Linnean Soc. 28, 65-82.
Rodriguez-Girons, M.A. & Santamaria, L. (2006). A new algorithm to calculate the nestedness temperature of presence-absence matrices. J. Biogeogr. 33, 924-935.
Stone, L. & Roberts, A. (1990). The checkerboard score and species distributions. Oecologia 85, 74-79.
Wright, D.H., Patterson, B.D., Mikkelson, G.M., Cutler, A. & Atmar, W. (1998). A comparative analysis of nested subset patterns of species composition. Oecologia 113, 1-20.